GO Annotation for the Immune System
Background
Immune System GO Curation: Tackling an 'Unmet Need'
The use of high-throughput technologies, such as gene expression, and systems biology as tools to investigate innate and adaptive immunity are gaining momentum. Gene-set enrichment analysis is often used to test for the statistically significant over-representation of particular pathways and functions. Such enrichment often relies on the availability of gene function, process and subcellular location annotations produced by the Gene Ontology Consortium. Currently the dissection of immune system responses is severely hampered by the lack of functional and pathway annotation of many of the key gene products involved.
Three members of the GO Consortium, based at Mouse Genome Informatics (MGI), European Bioinformatics Institute (EBI) and University College London (UCL), in collaboration with experts at Trinity College, Dublin (TCD), plan to tackle this annotation deficit with assistance from the immunological community and associated societies. Our objective is to provide comprehensive annotation of the thousands of mammalian genes that are required for the functioning of the immune system and the successful defense against pathogens using a well established controlled vocabulary, The Gene Ontology. This focus on immunological annotation follows a recent revision of the Gene Ontology which provides a more comprehensive representation of immunologically important processes (see Diehl et al., 2007, PMID:17267433 for further details).
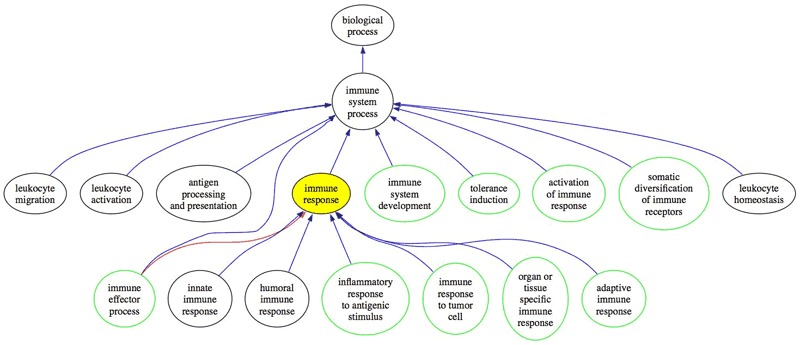
Figure illustrating the top level structure of the immune system processes in GO (Larger version)
Plan Of Action
- We have created a community annotation wiki for immunology intended to allow members of the community of experimental immunologists and other interested parties to comment on existing GO annotation and guide our continuing GO annotation efforts by pointing out missing experimentally established facts about particular genes involved in the immune system that are not currently represented by GO annotation.
- We have prepared a gene list of 3690 genes associated with the functioning of the immune system by combining and prioritizing several existing published lists of immune system genes. For further details, please see the Explanation of gene prioritization on the GO wiki.
How You Can Contribute
You can participate in this effort at many different levels.
- Contribute to the community annotation wiki for immunology. We are particularly keen to recruit immune gene and system process experts to review our annotations using the wiki. Users may simply supply us with the PubMed identifier of a key experimental publication for curation or provide more detailed information or commentary, such as pointing out any experimental data that might be controversial (these will be discussed with our immunology advisory panel). Information regarding genes in any species is welcome. We will review information provided through the wiki on a timely basis, and notify users of how their contributions have resulted in GO annotations.
- Suggest new GO terms related to immunology via the wiki.
All contributions are appreciated.
Data Dissemination
All immune GO annotations generated from this project will be freely available via the Gene Ontology Consortium website, Ensembl, and UniProtKB databases, and by the databases of the species-specific projects. These annotations will, de facto, be incorporated into Entrez Gene by the National Center for Biotechnology Information (NCBI).
A new dataset of annotated immune related genes, will also be created as a gene association file, the official GO Consortium format for exchange of data.
Additional Information
- GO immunology personnel: advisory board members, annotation managers, curators, and supporting societies
- Case Study: Using GO Annotations to Interpret Immunology Data