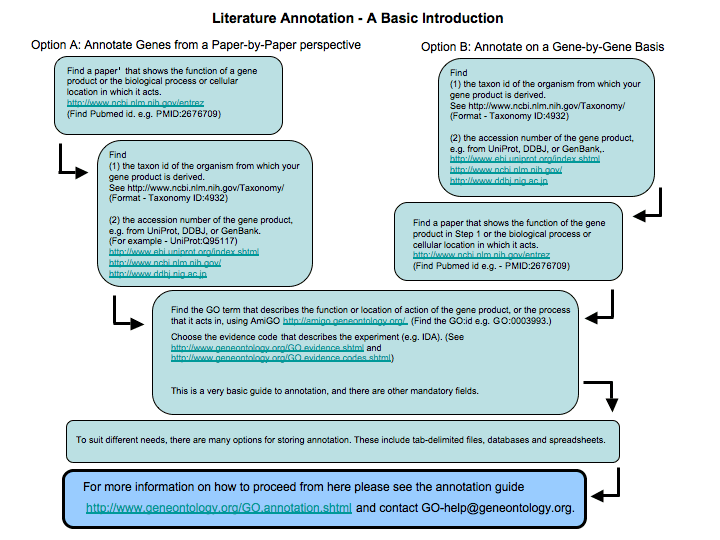
Annotation Guidelines
Annotation is the process of assigning GO terms to gene products. The annotation data in the GO database is contributed by members of the GO Consortium, and the Consortium is actively encouraging new groups to start contributing annotation. Annotations can be made from published literature where a curator reads and interprets the experiments and results presented in a paper or can be inferred automatically using sequence information or by key word mapping. Details on how to make automatic inferences can be found on the Electronic Annotation page. The GO annotation guide details more about the annotation process; other pages of interest may be the GO annotation conventions, the standard operating procedures used by some consortium members, and the GO annotation file format guide.
Literature Annotation
Literature annotation involves capturing published information about the exact function of a gene product as a GO annotations. To do this you must read the publications about the gene and write down all the information. This annotation is time-consuming but produces very high quality, species-specific annotation, and brings the information about the gene product into a format in which it can be used in high-throughput experiments. This is an extremely worthwhile process in the long term. It may be best carried out by people who know the function of the gene product, and the associated biology, in great detail; for example experimental scientists who are familiar with the published literature. If you are doing this, then you may like to write and suggest modifications to the ontology structure as well.
Annotation Conventions
This page contains guidelines which apply to all annotation methods and are particularly useful for manual literature-based annotation. Information on submitting annotations, database groups can be found in the GOC's annotation policy and the GO annotation standard operating procedures.
General recommendations
- A gene product can be annotated to zero or more nodes of each ontology.
- Annotation of a gene product to one ontology is independent of its annotation to other ontologies.
- Annotate gene products in each species database to the most detailed level in the ontology that correctly describes the biology of the gene product.
- Keep in mind that annotating to a term implies annotation to all parents via any path, so it is a good idea to check the parentage of a term before annotating (and request new terms or path corrections if necessary).
- Uncertain knowledge of where a gene product operates should be denoted by annotating it to two nodes, one of which can be a parent of the other. For instance, a yeast gene product known to be in the nucleolus, but also experimentally observed in the nucleus generally, can be annotated to both nucleolus and nucleus in the cell component ontology. Even though annotation to nucleolus alone implies that a gene product is also in the nucleus, annotate to both so as to explicitly indicate that it has been reported in the two locations. The two annotations may have the same or different supporting evidence. Similar reports of general and specific molecular function or biological process for a gene product could be handled the same way; for example, you may have direct experimental evidence (IDA) for DNA binding, but only a mutant phenotype (IMP) the more specific function term transcription factor activity and the process transcription. You also can annotate to multiple nodes that conflict with each other if there are conflicting claims in the literature.
- An individual gene product that is part of a complex can be annotated to terms that describe the action (function or process) of the complex. This practice is colloquially known as annotating 'to the potential of the complex', and is a way to capture information about what a complex does in the absence of database objects and identifiers representing complexes. For molecular function annotations, also see Using the Qualifier column below.
- A gene product should be annotated with terms reflecting its normal activity and location. A function, process, or localization (component) observed only in a mutant or disease state is therefore not usually included. In some circumstances, however, what is "normal" is a matter of perspective, depending on the organism being annotated and on the point of view of the annotator. For example, many viruses use host proteins to carry out viral processes. The host protein is then doing something abnormal from the perspective of the host, but completely normal from the perspective of the virus. GO annotators handle these cases by including two taxon IDs in the Taxon column of the gene association file; see annotating gene products that interact with other organisms for how to handle these cases.
- The evidence code No Data (ND) should be used as an indicator of curation status to denote gene products for which no relevant information could be found. It distinguishes gene products with no data available from those that have not yet been annotated. For more details on the code and its usage, please consult the ND evidence code documentation.
Core Information needed for a GO annotation
- Gene or Gene product Identifier (e.g. Q9ARH1)
- GO Term ID (e.g. GO:0004674, protein serine/threonine kinase)
- Reference ID (Pubmed:12374299 or GO_Ref:0000001)
- Evidence code (e.g. IDA or IMP)
Gene or Gene Product identifier (Database Objects)
Because a single gene may encode very different products with very different attributes, GO recommends associating GO terms with database objects representing gene products rather than genes. At present, however, many participating databases are unable to associate GO terms to gene products, and therefore use genes instead. If the database object is a gene, it is associated with all GO terms applicable to any of its products. See the annotation file format guide for more information.
Picking the appropriate GO term
The most difficult task in doing literature based annotations is in figuring out the correct GO term and the evidence code to use in the annotation. The GOC has identified some areas of the ontology that are more difficult to interpret and has come up with guidelines to help curators map the data/results presented in the paper to the right GO term. They are listed below.
Guidelines for annotating to Downstream processes
Guidelines for annotating to Binding and its child terms
Guidelines for using response_to and its child terms
Guidelines for using terms in the regulation branch
References and Evidence
Every annotation must be attributed to a source, which may be a literature reference, another database or a computational analysis.
The annotation must indicate what kind of evidence is found in the cited source to support the association between the gene product and the GO term. A simple controlled vocabulary of evidence codes is used to capture this; please see the GO evidence code documentation for more information on the meaning and use of the evidence codes.
Using the Qualifier column
The Qualifier column is used for flags that modify the interpretation of an annotation. Allowable values are NOT, contributes_to, and colocalizes_with.
NOT
NOT may be used with terms from any of the three ontologies.
NOT is used to make an explicit note that the gene product is not associated with the GO term. This is particularly important in cases where associating a GO term with a gene product should be avoided (but might otherwise be made, especially by an automated method). For example, if a protein has sequence similarity to an enzyme (whose activity is GO:nnnnnnn), but has been shown experimentally not to have the enzymatic activity, it can be annotated as NOT GO:nnnnnnn. (Note: in an email exchange from Sept. 2003 this phenomenon was referred to as "sequence dissimilarity.")
NOT can also be used when a cited reference explicitly says (e.g. "our favorite protein is not found in the nucleus"). Prefixing a GO ID with the string NOT allows annotators to state that a particular gene product is NOT associated with a particular GO term. This usage of NOT was introduced to allow curators to document conflicting claims in the literature.
Note that NOT is used when a GO term might otherwise be expected to apply to a gene product, but an experiment, sequence analysis, etc. proves otherwise. (It is not generally used for negative or inconclusive experimental results.)
colocalizes_with
colocalizes_with may be used only with cellular component terms.
Gene products that are transiently or peripherally associated with an organelle or complex may be annotated to the relevant cellular component term, using the colocalizes_with qualifier. This qualifier may also be used in cases where the resolution of an assay is not accurate enough to say that the gene product is a bona fide component member.
Example (from Schizosaccharomyces pombe):
Clp1p relocalizes from the nucleolus to the spindle and site of cell division; i.e. it is associated transiently with the spindle pole body and the contractile ring (evidence from GFP fusion). Clp1p is annotated to spindle pole body ; GO:0005816 and contractile ring ; GO:0005826, using the colocalizes_with qualifier in both cases.
contributes_to
contributes_to may be used only with molecular function terms.
As noted above, an individual gene product that is part of a complex can be annotated to terms that describe the function of the complex. Many such function annotations should use the qualifier contributes_to:
Annotating individual gene products according to attributes of a complex is especially useful for molecular function annotations in cases where a complex has an activity, but not all of the individual subunits do. (For example, there may be a known catalytic subunit and one or more additional subunits, or the activity may only be present when the complex is assembled.) Molecular function annotations of complex subunits that are not known to possess the activity of the complex must include the entry contributes_to in the Qualifier column. The contributes_to qualifier should not be used in biological process annotations. All gene products annotated using contributes_to must also be annotated to a cellular component term representing the complex that possesses the activity.
Annotations using contributes_to will often use the evidence code IC, but other codes may be used as well.
Note that contributes_to is not needed to annotate a catalytic subunit. Furthermore, contributes_to may be used for any non-catalytic subunit, whether the subunit is essential for the activity of the complex or not.
Examples
- Subunits of nuclear RNA polymerases: none of the individual subunits have RNA polymerase activity, yet all of these subunits are annotated to DNA-dependent RNA polymerase activity (with the contributes_to note), to capture the activity of the complex.
- ATP citrate lyase (ACL) in Arabidopsis: it is a heterooctamer, composed of two types of subunits, ACLA and ACLB in a A(4)B(4) stoichiometry. Neither of the subunits expressed alone give ACL activity, but co-expression results in ACL activity. Both subunits can be annotated to ATP citrate lyase activity.
- eIF2: has three subunits (alpha, beta, gamma); one binds GTP; one binds RNA; the whole complex binds the ribosome (all three subunits are required for ribosome binding). So one subunit is annotated to GTP binding and one to RNA binding without qualifiers, and all three are annotated to ribosome binding, with the contributes_to qualifier. And all three are annotated to the component term for eIF2 complex.
Sequence-based annotation
General principles for sequence IDs
- You must have stable identifiers for your objects.
- You must provide information on what the object is, e.g. a protein, nucleotide, EST, etc.. It doesn't matter if a nucleotide sequence is a gene, a genome, or an EST as long as it can be identified as such.
- If a sequence identifier has become obsolete, there must be a mechanism in place for tracking down the replacement.
- Your database must have an internal rule that object identifiers are never reused.
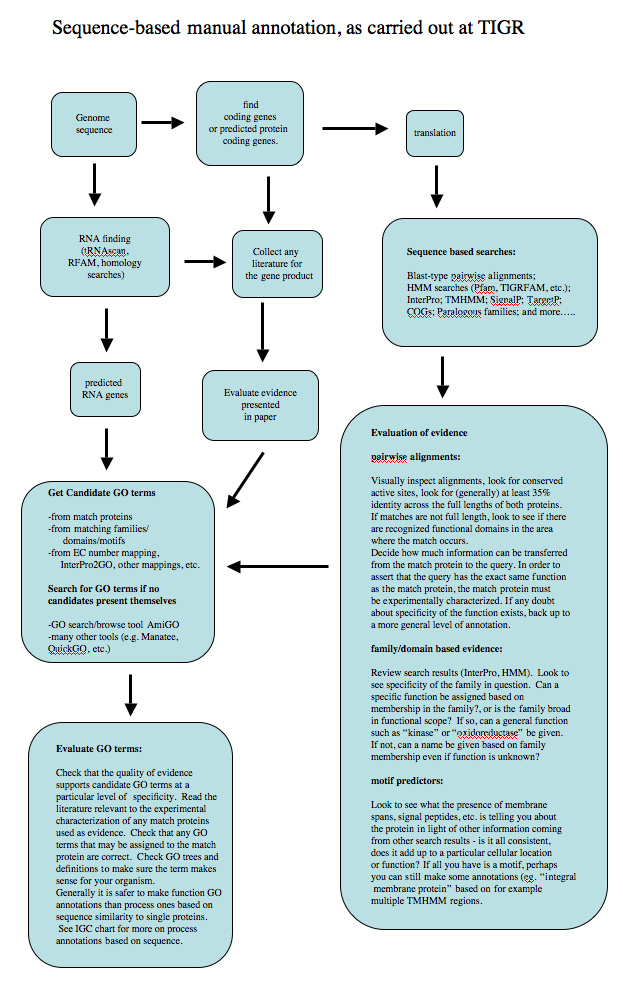
Annotation workflow
The following diagram shows the standard operating procedure for sequence-based (ISS evidence code) annotation used in the past at The Institute for Genomic Research (now JCVI).
Annotating gene products that interact with other organisms
The majority of gene products act within the organism that encoded them. However, sometimes gene products encoded by one organism can act on or in other organisms. For example, in obligate parasitic species (including viruses), almost all their gene products will be interacting with their host organism. Interactions may also be between organisms of the same species: for example, the proteins used by bacteria to adhere to one another to form a biofilm.
For annotating gene products involved in these multi-organism interactions, there are special terms in the biological process ontology, under multi-organism process, and in the cellular component ontology, under host. More specific information can be found in the biological process documentation on multi-organism processes and in the cellular component guidelines on host cell.
The species in the interaction are recorded in an annotation by using terms from this node and entering two taxon IDs in the Taxon column. The first taxon ID should be that of the species encoding the gene product, and the second should be the taxon of the other species in the interaction. Where the interaction is between organisms of the same species, both taxon IDs should be the same. The taxon column of the annotation file is described in more detail in the annotation file format guide.
An additional taxon ID should not be added in cases where the annotation is based on sequence or structural similarity.
Nomenclature Conventions
The terms 'symbiont' and 'host' may carry connotations of the nature of the interaction between two organisms, but in the Gene Ontology, they are used solely to differentiate between organisms on the basis of their size. The word symbiont is used to refer to the smaller organism in a symbiotic interaction; the larger organism is called the host. If the two organisms are the same size, the term will be contain other organism. Note that parasites and pathogens are also referred to as 'symbionts', as symbiosis encompasses parasitism, commensalism and mutualism.
Requesting new terms in the multi-organism process node
Like the rest of GO, the multi-organism process node is not complete, and you will probably have to request some new terms when annotating your gene products. These should be submitted via the GO curator requests tracker in the usual way. Here are a few points to bear in mind when requesting new terms, and annotating using this node:
- A term name should make the direction of the interaction clear. An example of this is given below; induction of nodule morphogenesis in host would be used to annotate the symbiont gene product, while induction of nodule morphogenesis by symbiont is used to annotate the host genes. Both processes would be children of a common term nodulation.
- If your gene product affects a 'normal' host process, you should always request a new term in the MOP node, rather than just annotating directly to the term in the 'normal' ontology. So for example, if your bacterial gene product regulates the ethylene-mediated signaling pathway in plants, rather than using dual taxon to annotate to regulation of ethylene mediated signaling pathway ; GO:0010104, you should instead request a new term regulation of ethylene mediated signaling pathway in host.
- Where an organism subverts a 'normal' biological process, e.g. the transcription of viral DNA by host transcription machinery, host proteins should not be annotated to a 'symbiont' term like transcription of symbiont DNA. This is because this would be considered considered a pathological process, i.e. not 'normal' for the host.
Example: Performing a process with another organism
Nod factor export proteins transfer nod factors out of the purple bacterium Sinorhizobium meliloti into the surrounding soil. Here they are detected by LysM nod factor receptor kinases in Medicago truncatula roots and initiate the process of nodulation.
Annotation of Nod factor export ATP-binding protein I from S. meliloti
suggest a new term induction of nodule morphogenesis in host
nodulation ; GO:0009877
[p] induction of nodule morphogenesis in host ; GO:00new01
Sinorhizobium meliloti taxonomy ID: 382
Medicago truncatula taxonomy ID: 3880
protein name: Nod factor export ATP-binding protein I
GO term: induction of nodule morphogenesis in host ; GO:00new01
taxon column: taxon:382|taxon:3880
Annotation of LysM receptor kinase LYK3 precursor from M. truncatula
suggest a new term induction of nodule morphogenesis by symbiont
nodulation ; GO:0009877
[p] induction of nodule morphogenesis by symbiont ; GO:00new02
Medicago truncatula taxonomy ID: 3880
Sinorhizobium meliloti taxonomy ID: 382
protein name: LysM receptor kinase LYK3 precursor
GO term: induction of nodule morphogenesis by symbiont ; GO:00new02
taxon column: taxon:3880|taxon:382
Example: Performing a process in more than one species
The protein cardiotoxin from the southern Indonesian spitting cobra Naja sputatrix kills mammalian cells by cytolysis when it enters the host cell cytoplasm.
Annotation of cardiotoxin precursor, from N. sputatrix
use the GO terms cytolysis of cells of another organism ; GO:0051715 and host cell cytoplasm ; GO:0030430
Naja sputatrix taxonomy ID: 33626
Mammalia taxonomy ID: 40674
protein name: cardiotoxin precursor
GO term: cytolysis of cells of another organism ; GO:0051715
taxon column: taxon:33626|taxon:40674
protein name: cardiotoxin precursor
GO term: host cell cytoplasm ; GO:0030430
taxon column: taxon:33626|taxon:40674
Example: Regulating a process in another organism
Mosquito saliva contains D7 proteins, which bind biogenic amines in order to suppress hemostasis in humans.
Annotation of D7 protein long form, from A. gambiae
suggest a new term negative regulation of hemostasis in host
evasion of host defense response ; GO:0030682
[i] negative regulation of hemostasis in host ; GO:00new03
Anopheles gambiae taxonomy ID: 7165
Homo sapiens taxonomy ID: 9606
protein name: D7 protein long form
GO term: negative regulation of hemostasis in host ; GO:00new03
taxon column: taxon:7165|taxon:9606
Annotation mailing list
All Consortium annotators should subscribe to the GO discussion mailing list, which provides a forum for the discussion of annotations and specific use questions. Subscription details and archived posts are available on the annotation mailing list information page.
Specific annotation queries can be submitted to the GO annotation tracker at SourceForge. For general queries about annotation not answered by this page, please email the GO helpdesk.
GO Annotation Resources
For more information on annotation, please see the following resources: