Annotation Basics
Annotation is the process of assigning GO terms to gene products. The annotation data in the GO database is contributed by members of the GO Consortium, and the Consortium is actively encouraging new groups to start contributing annotation. Annotations can be made from published literature where a curator reads and interprets the experiments and results presented in a paper or can be inferred automatically using sequence information or by key word mapping. Details on how to make automatic inferences can be found on the Electronic Annotation page. The GO annotation guide details more about the annotation process; other pages of interest may be the GO annotation conventions, the standard operating procedures used by some consortium members, and the GO annotation file format guide.
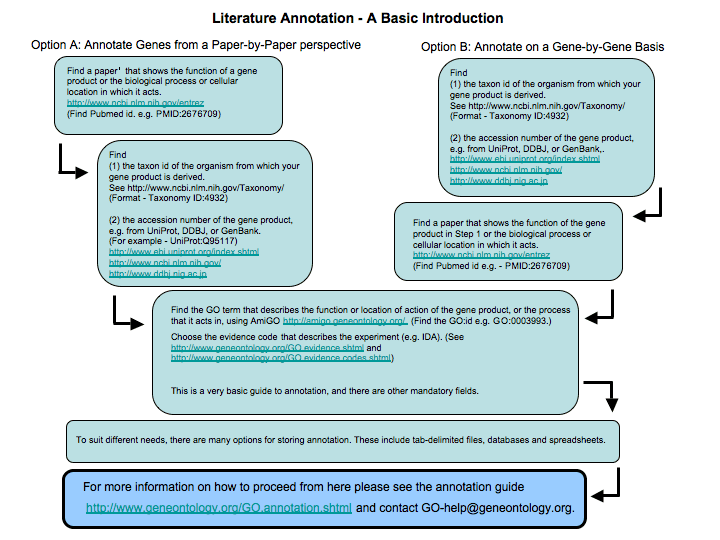
Literature Annotation
Literature annotation involves capturing published information about the exact function of a gene product as a GO annotations. To do this you must read the publications about the gene and write down all the information. This annotation is time-consuming but produces very high quality, species-specific annotation, and brings the information about the gene product into a format in which it can be used in high-throughput experiments. This is an extremely worthwhile process in the long term. It may be best carried out by people who know the function of the gene product, and the associated biology, in great detail; for example experimental scientists who are familiar with the published literature. If you are doing this, then you may like to write and suggest modifications to the ontology structure as well.
Annotation Conventions
This page contains guidelines which apply to all annotation methods. Information on submitting annotations, database groups can be found in the GOC's annotation policy and the GO annotation standard operating procedures.
General recommendations
- A gene product can be annotated to zero or more nodes of each ontology.
- Annotation of a gene product to one ontology is independent of its annotation to other ontologies.
- Annotate gene products in each species database to the most detailed level in the ontology that correctly describes the biology of the gene product.
- Keep in mind that annotating to a term implies annotation to all parents via any path, so it is a good idea to check the parentage of a term before annotating (and request new terms or path corrections if necessary).
- Uncertain knowledge of where a gene product operates should be denoted by annotating it to two nodes, one of which can be a parent of the other. For instance, a yeast gene product known to be in the nucleolus, but also experimentally observed in the nucleus generally, can be annotated to both nucleolus and nucleus in the cell component ontology. Even though annotation to nucleolus alone implies that a gene product is also in the nucleus, annotate to both so as to explicitly indicate that it has been reported in the two locations. The two annotations may have the same or different supporting evidence. Similar reports of general and specific molecular function or biological process for a gene product could be handled the same way; for example, you may have direct experimental evidence (IDA) for DNA binding, but only a mutant phenotype (IMP) the more specific function term transcription factor activity and the process transcription. You also can annotate to multiple nodes that conflict with each other if there are conflicting claims in the literature.
- An individual gene product that is part of a complex can be annotated to terms that describe the action (function or process) of the complex. This practice is colloquially known as annotating 'to the potential of the complex', and is a way to capture information about what a complex does in the absence of database objects and identifiers representing complexes. For molecular function annotations, also see Using the Qualifier column below.
- A gene product should be annotated with terms reflecting its normal activity and location. A function, process, or localization (component) observed only in a mutant or disease state is therefore not usually included. In some circumstances, however, what is "normal" is a matter of perspective, depending on the organism being annotated and on the point of view of the annotator. For example, many viruses use host proteins to carry out viral processes. The host protein is then doing something abnormal from the perspective of the host, but completely normal from the perspective of the virus. GO annotators handle these cases by including two taxon IDs in the Taxon column of the gene association file; see annotating gene products that interact with other organisms for how to handle these cases.
- The evidence code No Data (ND) should be used as an indicator of curation status to denote gene products for which no relevant information could be found. It distinguishes gene products with no data available from those that have not yet been annotated. For more details on the code and its usage, please consult the ND evidence code documentation.
Core Information needed for a GO annotation
- Gene or Gene product Identifier (e.g. Q9ARH1)
- GO Term ID (e.g. GO:0004674, protein serine/threonine kinase)
- Reference ID (Pubmed:12374299 or GO_Ref:0000001)
- Evidence code (e.g. IDA or IMP)
Gene or Gene Product identifier (Database Objects)
Because a single gene may encode very different products with very different attributes, GO recommends associating GO terms with database objects representing gene products rather than genes. At present, however, many participating databases are unable to associate GO terms to gene products, and therefore use genes instead. If the database object is a gene, it is associated with all GO terms applicable to any of its products. See the annotation file format guide for more information.
Picking the appropriate GO term
The most difficult task in making an annotation is in
figuring out the correct GO term and the evidence code to use in the
annotation.
The GOC has identified some areas of the ontology that are more
difficult to interpret and has come up with a
References and Evidence
Every annotation must be attributed to a source, which may be a literature reference, another database or a computational analysis.
The annotation must indicate what kind of evidence is found in the cited source to support the association between the gene product and the GO term. A simple controlled vocabulary of evidence codes is used to capture this; please see the GO evidence code documentation for more information on the meaning and use of the evidence codes.
Using the Qualifier column
The Qualifier column is used for flags that modify the interpretation of an annotation. Allowable values are NOT, contributes_to, and colocalizes_with. More information can be found in the documentation on Qualifiers
Sequence-based annotation
General principles for sequence IDs
- You must have stable identifiers for your objects.
- You must provide information on what the object is, e.g. a protein, nucleotide, EST, etc.. It doesn't matter if a nucleotide sequence is a gene, a genome, or an EST as long as it can be identified as such.
- If a sequence identifier has become obsolete, there must be a mechanism in place for tracking down the replacement.
- Your database must have an internal rule that object identifiers are never reused.
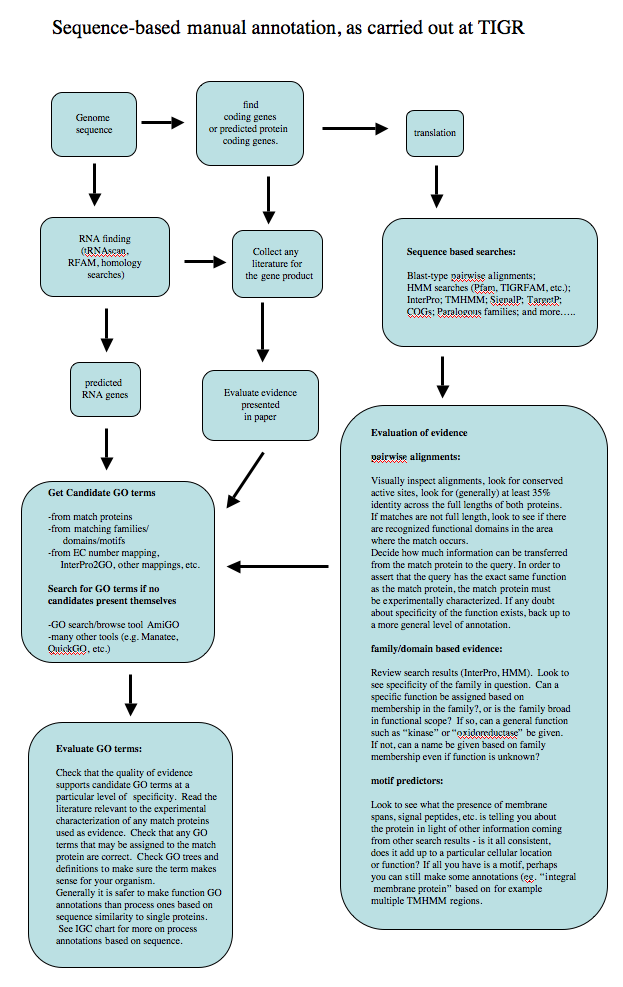
Annotation workflow
The following diagram shows the standard operating procedure for sequence-based (ISS evidence code) annotation used in the past at The Institute for Genomic Research (now JCVI).
Annotation mailing list
All Consortium annotators should subscribe to the GO discussion mailing list, which provides a forum for the discussion of annotations and specific use questions. Subscription details and archived posts are available on the annotation mailing list information page.
Specific annotation queries can be submitted to the GO annotation tracker at SourceForge. For general queries about annotation not answered by this page, please email the GO helpdesk.
GO Annotation Resources
For more information on annotation, please see the following resources: