Browsing and searching GO and its annotations
The Gene Ontology is a controlled vocabulary of terms to describe gene product characteristics in the domains of localization and function. Databases using GO terms to annotate their genes and gene products can submit their annotations to the GO consortium where they are made freely available for other users to download and utilize. This tutorial will show you how to browse and search the Gene Ontology and the annotations made using its terms.
GO Browsers
The first part of this tutorial uses the AmiGO Browser (http://www.godatabase.org/), developed by the Gene Ontology Consortium, but there are also many other GO browsers (see http://www.geneontology.org/GO.tools.html) developed by outside groups that can be used.
Browsing GO using AmiGO
Open AmiGO at http://www.godatabase.org/
This will display the Gene Ontology graph with the three ontologies: biological process, cellular component and molecular function, as well as the three nodes of obsolete terms, containing terms that have been removed from the active ontologies.
|
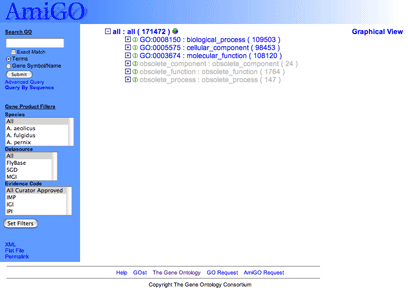
|
You can navigate the tree by clicking on the  and and  icons at the start of each line. icons at the start of each line.
 expands a node, showing all the children of the term. expands a node, showing all the children of the term.
 closes the node, hiding the children. closes the node, hiding the children.
 means that the term on that line has no children. means that the term on that line has no children.
Greyed out terms are obsolete, meaning that they are deprecated and should no longer be used.
|
Each instance of a term gets one horizontal line.

The  and
and  icons represent the relationship of the term to its parent, either "is a" and "part of" the parent term.
icons represent the relationship of the term to its parent, either "is a" and "part of" the parent term.
The GO term identifier and term name can be clicked to get a more detailed view of the term, including the definition and all genes and gene products annotated to the term. Mousing over the term brings up a floating box showing the term definition.
Following the term ID and name is a number in parentheses. This is the total number of genes manually annotated to this term and its children. Electronic annotations (evidence code IEA) are not shown for two reasons: there are large numbers of these annotations, and they are deemed lower quality as they have not been checked by a human.
Terms may be followed by the  icon. Clicking this icon will bring you to a pie chart which displays the percentage of gene products annotated to each term below that selected.
icon. Clicking this icon will bring you to a pie chart which displays the percentage of gene products annotated to each term below that selected.
Open the node biological process, then open physiological process, then secretion
How many of the children of secretion have no children of their own?
How many annotations (non-IEA) does acid secretion have to it?
What relationship does regulation of secretion have to its parent secretion?
Clicking on a term name or ID opens the term detail page.
|
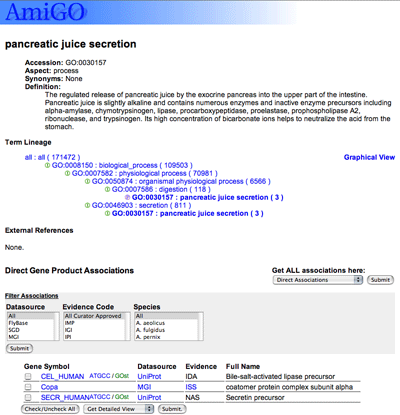
|
The term detail page shows all the information available about the term: the term name and ID, any synonyms it might have, the term definition, its position in the GO structure, references to external databases, and the gene products associated with that term. Note that children of the selected term are not shown.
You can click on "Graphical view" for an alternate representation of the tree structure.
|
Open the node protein secretion and click on the term cytokine secretion. It will open in a new window.
What are the parents of cytokine secretion and what relationships does it have to them?
Click on the parent term protein secretion to retrieve the information about it.
Following the tree view is a list of external references, which are links to equivalent concepts in other databases (eg. EC numbers, MIPS functional classifications) or objects which have been given GO annotation (eg. sequence features or protein families). Click on the  icon to display the database references; some are linked directly to the external databases.
icon to display the database references; some are linked directly to the external databases.
For more info on the contributing databases see the Gene Ontology website indices to other classification systems (http://www.geneontology.org/GO.indices.html) and acknowledgements page (http://www.geneontology.org/GO.acknowledgements.html).
Beneath the term information are the annotations, the genes or gene products assigned the selected term.
|
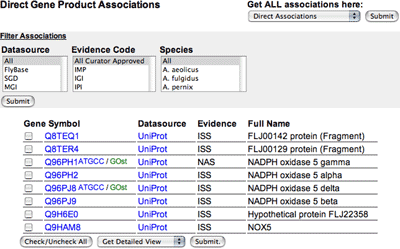
|
The first column is the gene or gene product identifier; clicking on the name will take you to the AmiGO gene product detail page, which shows the information held in the GO database about that gene product, including all its GO annotations and the peptide sequence (if available).
The second column is the data source that submitted the annotation (e.g. FlyBase, SGD, UniProt), and clicking on this takes you to the detail page at the source's website.
The third column is the evidence code for the association; when underlined, clicking on the evidence code brings up the source reference used to make the association.
The final column has the full name of the gene product where available.
|
You can choose to view the annotations to the term itself or to the term and its children.
Go back to the cytokine secretion term detail page. From the drop-down menu on the right, choose get "All associations with terms" and click submit.
How many gene products are annotated directly to cytokine secretion and how many are annotated to its child terms?
Which databases have made these annotations?
Note the filtering menus in the light grey box. You can also choose to filter annotations by the database that supplied them, by the evidence code used in the annotation and by species.
Change the view so that gene associations from mouse (M. musculus) are displayed.
Which databases have submitted mouse gene annotations?
Shut the popup window and return to the main page.
Searching with AmiGO
|
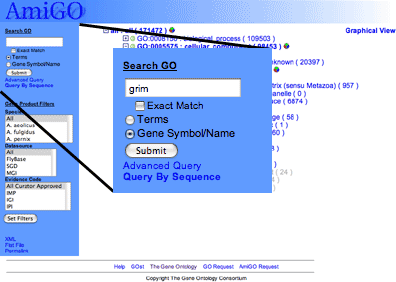
|
At the top of the page there is a search box. GO terms or associated gene products can be searched by checking the "terms" or "gene products" boxes respectively.
|
Perform a search for gene products containing the text 'grim'.
|
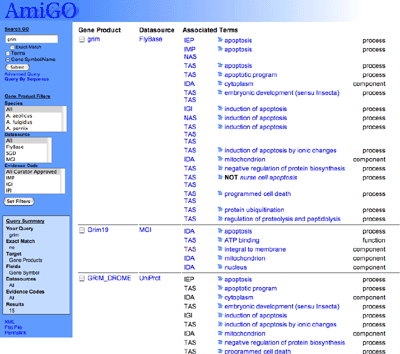
|
The results list displays all gene products containing the text "grim", the name of the external database which the gene product is from, all GO terms to which the product has been associated and the types of evidence linking the gene product to that term, and the aspect - function, process, component - that the GO term describes.
|
How many GO terms is the FlyBase gene product "Buffy" associated with?
Do a search for your favourite gene product.
Does this gene product already have a GO association?
Perform a search for GO terms containing the text rough endoplasmic reticulum.
|
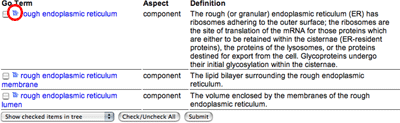
|
Each row of the results table contains one GO term, its aspect and the definition, where available. Clicking the term will bring up the detailed view. Clicking the icon to the left of the term name (ringed in red) will show your term placed in the GO tree.
|
How many gene products are annotated to the term rough endoplasmic reticulum membrane ; GO:0030867?
Pie Charts
Return to the AmiGO main page (http://www.godatabase.org) and expand the term biological process to view its children. Click the pie icon that now appears next to biological process to view the annotations as a pie chart.
What term makes up the biggest slice of the biological process annotations?
Close the pie chart window and use the filtering controls under the search box on the left to view associations from SGD (Saccharomyces Genome Database). Click "Set filters" and then view a pie chart of the distribution of associations under biological process.
Has the distribution changed at all?
GOst
GOst is the Gene Ontology Blast server, which allows you to blast a protein sequence against all gene products that have a GO annotation.
Return to the main AmiGO page and click "GOst search" (at the bottom of the page). Enter the UniProt accession number Q61337 into the top box and click submit.
What are the results? What GO terms are associated with them?
Useful Links
The Gene Ontology Consortium website, http://www.geneontology.org/
QuickGO, http://www.ebi.ac.uk/ego/ - alternative GO browser maintained and run by the European Bioinformatics Institute

 and
and  icons at the start of each line.
icons at the start of each line.
 expands a node, showing all the children of the term.
expands a node, showing all the children of the term.
 closes the node, hiding the children.
closes the node, hiding the children.
 means that the term on that line has no children.
means that the term on that line has no children.

 and
and  icons represent the relationship of the term to its parent, either "is a" and "part of" the parent term.
icons represent the relationship of the term to its parent, either "is a" and "part of" the parent term.
 icon. Clicking this icon will bring you to a pie chart which displays the percentage of gene products annotated to each term below that selected.
icon. Clicking this icon will bring you to a pie chart which displays the percentage of gene products annotated to each term below that selected.




